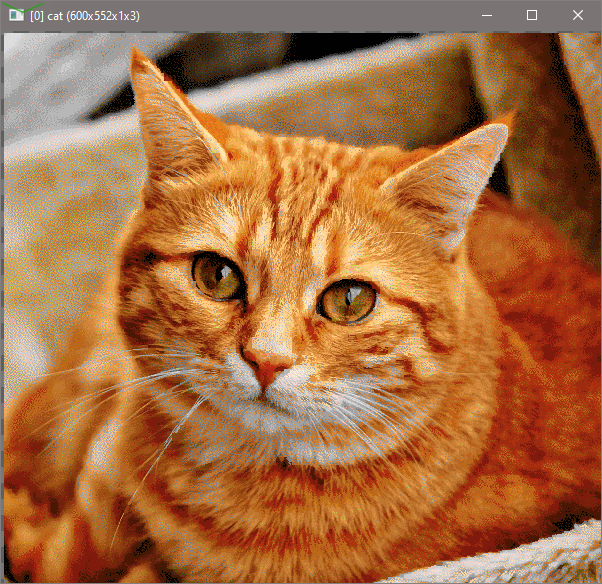
I’m working on a new version of Stitch. It will be faster. It is very hard to code up too.
For picture of what Stitch looks like:
Here’s what the current code looks like:
Code to New Stitch
#@cli new_rep_stitch: offset,thickness,modulo,interpolation,boundary,0<sublevel,randomize_stitching,
#@cli : Stitches images.
new_rep_stitch:
skip ${3=0},${8=}
# offset = $1(%)
# thickness = $2(%)
# modulo = $3 >= 0
# interpolation = $4 > 0
# boundary = $5 >= 0
# sublevel = abs($6) + 1
# random_stitching = $7 => { 0=Do-Not-Randomize | 1=Use Randomization On Strip | 2=Use Randomization On Offset | 3= Use Randomization on Offset and Strip }
# seed_a = $8 => For offset
# seed_b = $9 => For each strip
# angles = $9.....$inf
if ${is_percent\ $1} use_perc_off=1
else
use_perc_off,offset=0,{abs($1)*$sublevel}
if !$offset error abs("$1")!=0==F fi
fi
if ${is_percent\ $2} use_perc_thk=1
else
use_perc_thk,thickness=0,{abs($2)*$sublevel}
if !$thickness error abs("$2")!=0==F fi
fi
modulo,interpolation,boundary,sublevel,random_stitch_mode,seed_off,seed_strip,generate_out_mode,timg,max_img_ind={abs([${3-5}])},{abs($6)+1},{$7%4},${8-9},0,{$!},{$!-1}
if $modulo if $modulo<2 error "$3">1==F fi fi
+rep_find_nonduplicate_angles ${10--1} :: rad_ang
num_angs={w#-1}
if $!>2
eval "
diff=0;
const ti=$timg;
initial_dimensions=[w#0,h#0,d#0];
for(p=1,p<ti,++p,
if([w#p,h#p,d#p]!=initial_dimensions,
diff=1;
break();
);
);
diff;"
generate_out_mode=${}
fi
if !$generate_out_mode
r[0--2] {w#0*$sublevel},{h#0*$sublevel},100%,100%,1
length={norm(w#0,h#0)/2}
if $use_perc_off offset={$length*$1} fi
if $use_perc_thk thickness={int($length*$2)} fi
if $offset&&$thickness
+f. "begin(
const thickness=$thickness;
const cx=(w#0-1)/2;
const cy=(h#0-1)/2;
const pt_1=atan2(cy,-cx);
const pt_0=atan2(cy,cx);
count_bars(v)=int(v/thickness)+1;
);
if(inrange(i,pt_0,pt_1,1,0)
,count_bars(1/sin(i)*cy)
,count_bars(1/cos(i)*cx)
);"
:: number_of_bars
if $random_stitch_mode>1
#Insert multiple randomization here#
error development
else
{$modulo?min($modulo,iM#-1):iM#-1},1,1,2
eval "
if(narg($seed_off),srand($seed_off););
const length=w#-1;
I(#-1)=[u(-1,1)*$offset,length];
for(p=1,p<length,++p,
a=u(-1,1)*$offset;
b=u(-1,1)*$offset;
if(int(u(0,2)),I(#-1,p)=[b,a],I(#-1,p)=[a,b]);
);"
fi
f[0-$max_img_ind] "
begin(
if(narg($seed_strip),srand($seed_strip););
const max_x=w-1;
const max_y=h-1;
const cx=max_x/2;
const cy=max_y/2;
const rs_mode=$random_stitch_mode;
const num_angs=$num_angs;
const length=w#-1;
const sub_length=length-1;
const thickness=$thickness;
const half_thickness=thickness>>1;
nbars=crop(#$number_of_bars);
angs=crop(#$rad_ang);
cos_ang=cos(angs);
sin_ang=sin(angs);
rot_x(a,b,c)=a*cos_ang[c]-b*sin_ang[c];
rot_y(a,b,c)=a*sin_ang[c]+b*cos_ang[c];
if(rs_mode<2,
pos_off=crop(#-1,0,0,0,0,1,w#-1,1,1);
neg_off=crop(#-1,0,0,0,1,1,w#-1,1,1);
rs_mode?(
neg_v=expr('>begin(if(narg($seed_strip),srand($seed_strip-1);););int(u(0,$num_angs+1));',length);
pos_v=expr('>begin(if(narg($seed_strip),srand($seed_strip+1);););int(u(0,$num_angs+1));',length);
neg_v=reverse(neg_v);
middle=expr('if(narg($seed_strip),srand($seed_strip););int(u(0,$num_angs+1));',1);
rand_v=[neg_v,middle,pos_v];
off_out(v)=rand_v[v+nbars[k]];
):(
off_out(v)=v%2;
);
strip_out(v)=v%num_angs;
);
);
xx=x-cx;
yy=y-cy;
ind=0;
repeat(num_angs,k,
ind+=off_out(floor((rot_y(xx,yy,k)+half_thickness)/thickness));
);
ind%=num_angs;
"
fi
fi
# repeat $!-1 l[$>,-1] {
#
# }
# done
#@cli rep_find_nonduplicate_angles: angle_0,angle_1,....,angle_n
#@cli : Return all non-duplicate angles in radian form.
+rep_find_nonduplicate_angles:
($*) (0)
eval[-2] "res_to_0_180(angle)=(angle%180)/180;
if(x,
v=res_to_0_180(i);
not_found=1;
for(p=0,p<da_size(#-1),++p,
if(i[#-1,p]==v,
not_found=0;
break();
);
);
if(not_found,da_push(#-1,v););
,
da_push(#-1,res_to_0_180(i));
);
end(resize(#-1,1,da_size(#-1),1,1,0););"
*. {pi} rotate. -90 rm..
The idea is to use only one image to do what the old Stitch does.
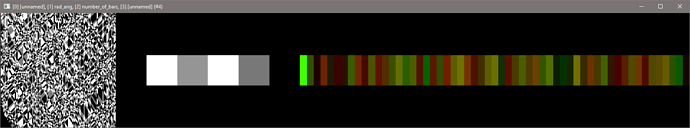
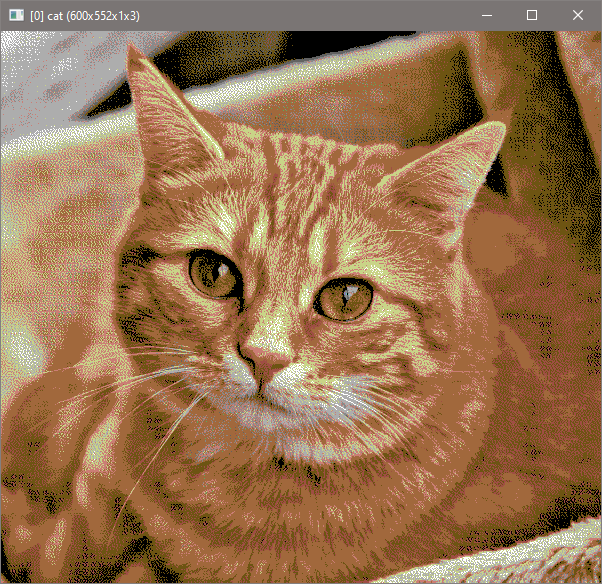
Here’s the output:
The last image are offset values. The first image will be used to generate the stitch output.
The timing shows real potential in being a really fast filter.